5nm transistors; reducing deep learning computations; biological NOR gates.
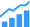
Silicon nanosheet-based builds 5nm transistor
To enable the manufacturing of 5nm chips, IBM, GLOBALFOUNDRIES, Samsung, and equipment suppliers have developed what they say is an industry-first process to build 5nm silicon nanosheet transistors. This development comes less than two years since developing a 7nm test node chip with 20 billion transistors. Now, they’ve paved the way for 30 billion switches on a fingernail-sized chip.
Pictured: a scan of IBM Research Alliance’s 5nm transistor, built using an industry-first process to stack silicon nanosheets as the device structure – achieving a scale of 30 billion switches on a fingernail-sized chip that will deliver significant power and performance enhancements over today’s state-of-the-art 10nm chips. (Source: IBM)
The team reported the resulting increase in performance should help accelerate cognitive computing, IoT, and other data-intensive applications delivered in the cloud; the power savings could also mean that the batteries in smartphones and other mobile products could last two to three times longer than today’s devices, before needing to be charged.
The researchers working as part of the IBM-led Research Alliance at the SUNY Polytechnic Institute Colleges of Nanoscale Science and Engineering’s NanoTech Complex in Albany, NY explained that they achieved the breakthrough by using stacks of silicon nanosheets as the device structure of the transistor, instead of the standard FinFET architecture, which is the blueprint for the semiconductor industry up through 7nm node technology.
Further, the researchers pointed out that compared to the leading edge 10nm technology, this nanosheet-based 5nm technology can deliver 40 percent performance enhancement at fixed power, or 75 percent power savings at matched performance.
Interestingly, this approach uses Extreme Ultraviolet (EUV) lithography, just as was used for the 7nm test node, and its 20 billion transistors was applied to the nanosheet transistor architecture. Using EUV lithography, the width of the nanosheets can be adjusted continuously, all within a single manufacturing process or chip design. This adjustability permits the fine-tuning of performance and power for specific circuits – something not possible with today’s FinFET transistor architecture production, which is limited by its current-carrying fin height. Therefore, while FinFET chips can scale to 5nm, simply reducing the amount of space between fins does not provide increased current flow for additional performance, the team added.
Reducing deep learning computations
To eliminate more than 95% of the computations that deep learning requires, Rice University researchers have adapted a widely used technique for rapid data lookup to slash the amount of computation — and thus energy and time — required.
Anshumali Shrivastava, lead researcher and assistant professor of computer science at Rice said, “This applies to any deep-learning architecture, and the technique scales sublinearly, which means that the larger the deep neural network to which this is applied, the more the savings in computations there will be.”
The team said this work addresses one of the biggest issues facing tech giants like Google, Facebook and Microsoft as they race to build, train and deploy massive deep-learning networks for a growing body of products as diverse as self-driving cars, language translators and intelligent replies to emails.
Shrivastava and Rice graduate student Ryan Spring have shown that techniques from “hashing,” a tried-and-true data-indexing method, can be adapted to dramatically reduce the computational overhead for deep learning. Hashing involves the use of smart hash functions that convert data into manageable small numbers called hashes. The hashes are stored in tables that work much like the index in a printed book.
The approach blends two techniques — a clever variant of locality-sensitive hashing and sparse backpropagation — to reduce computational requirements without significant loss of accuracy. For example, they said, in small-scale tests they found they could reduce computation by as much as 95 percent and still be within 1 percent of the accuracy obtained with standard approaches.
Circuits built in living cells
In order to harness the potential of cells as living computers that can respond to disease, efficiently produce biofuels or develop plant-based chemicals, University of Washington synthetic biology researchers have demonstrated a new method for digital information processing in living cells, analogous to the logic gates used in electric circuits — so they don’t have to wait for evolution to craft their desired cellular system.
They reminded that living cells must constantly process information to keep track of the changing world around them and arrive at an appropriate response. And through billions of years of trial and error, evolution has arrived at a mode of information processing at the cellular level. In the microchips that run our computers, information processing capabilities reduce data to unambiguous zeros and ones. In cells, it’s not that simple. DNA, proteins, lipids and sugars are arranged in complex and compartmentalized structures.
The team built a set of synthetic genes that function in cells like NOR gates, commonly used in electronics, which each take two inputs and only pass on a positive signal if both inputs are negative. NOR gates are functionally complete, meaning one can assemble them in different arrangements to make any kind of information processing circuit. And they did all of this using DNA instead of silicon and solder, and inside yeast cells instead of at an electronics workbench. The circuits the researchers built are the largest ever published to date in eurkaryotic cells, which, like human cells, contain a nucleus and other structures that enable complex behaviors.
Cells could potentially be reprogrammed to undergo new developmental pathways, to regrow organs or to develop entirely new ones. In such developing tissues, cells have to make complex digital decisions about what genes to express and when, and the new technology could be used to control that process, they noted.
While implementing simple programs in cells will never rival the speed or accuracy of computation in silicon, genetic programs can interact with the cell’s environment directly, explained UW electrical engineering professor Eric Klavins. “For example, reprogrammed cells in a patient could make targeted, therapeutic decisions in the most relevant tissues, obviating the need for complex diagnostics and broad spectrum approaches to treatment.”
Leave a Reply